Blog
Multi-Model in Action: Using RedisGraph and RediSearch together
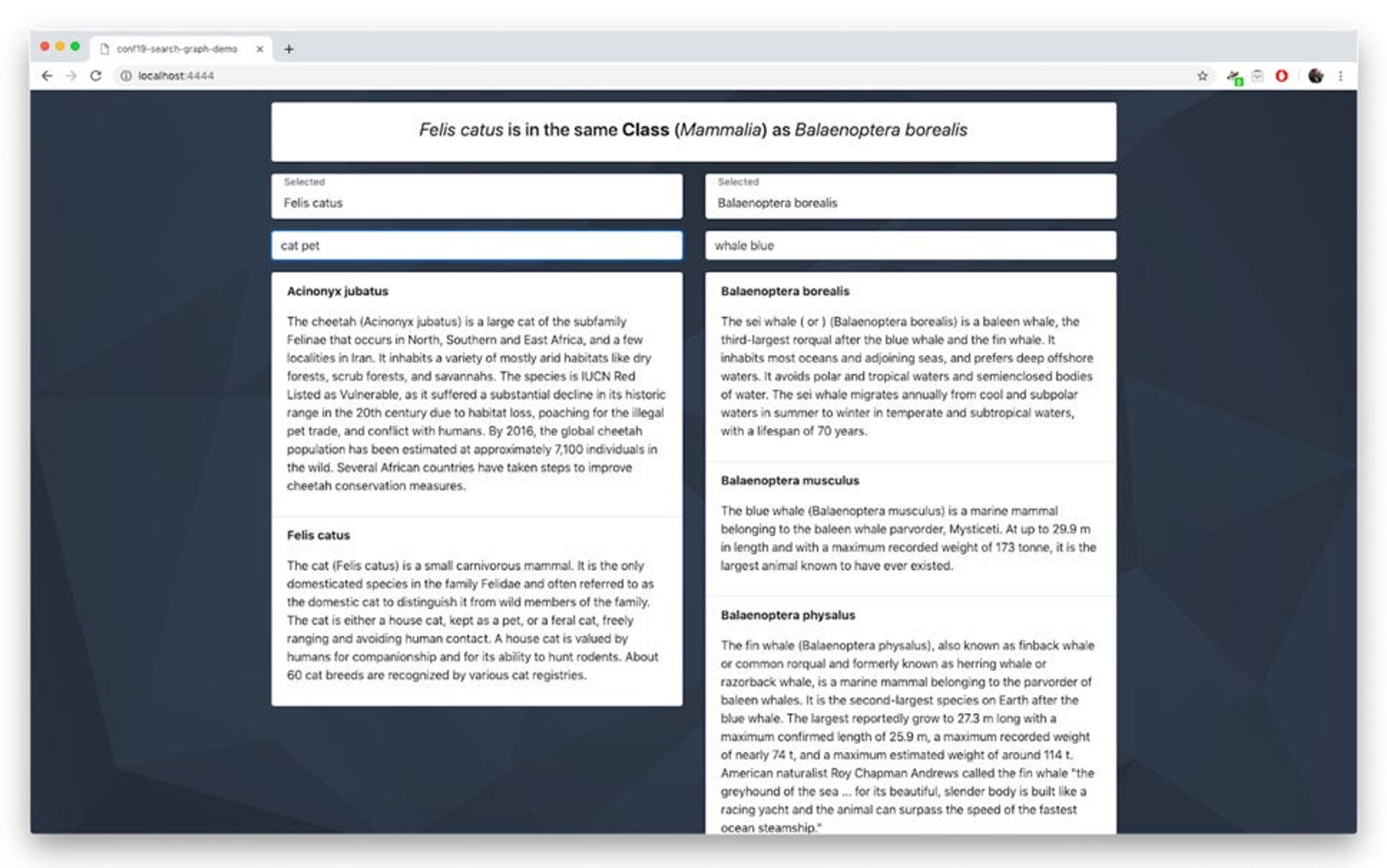
At RedisConf19, I demoed a solution for running full-text RediSearch over nodes in RedisGraph. The days leading up to the event are a bit of a fog, but now, a few weeks later, I realized we should explain more about how we did this and release the source code.
In this demo, I showed a little interface that allows you to search for animals and see how they are related via the biological classification system (kingdom, phylum, class, order and so on). The full-text portion was based on the first English paragraph from wikipedia. So, as an example, let’s say you searched for “cat house pet” (Domestic Cat, Felis catus) and “baleen blue” (Blue Whale, Balaenoptera musculus), you would see that they are both just mammals, while if you searched for “cat house pet” and “snow central asia cat” (Snow Leopard, Panthera uncia), you would see that they both belong to the same family, Felidae.
This demo project was surprisingly easy, but I should point out that the integration between RediSearch and RedisGraph is still early and not ready for production at the time of writing. I encourage you to figure out if this approach could meet your needs, with the understanding that the integration between RediSearch and RedisGraph will mature and expand in the coming months.
In the meantime, let’s talk about the components of the demo. The first thing to accomplish is to build both RediSearch and RedisGraph based on the correct branches in the repository. For RediSearch, that’s just the current master branch, while for RedisGraph you’ll need the redisconf branch. If you’d like to replicate the solution for your own needs, you’ll want to build both modules from source. The website for RedisGraph and RediSearch each have detailed instructions on how to do this, which takes a bit of time, but isn’t hard.
In your redis.conf file, you’ll need to ensure that RedisGraph is loaded before RediSearch. To do this, go to the redis.conf file and in the modules section, put the loadmodule directive for RediSearch before that of RedisGraph. This way, when RedisGraph loads, it will detect the presence of the search module. After you’ve finished editing redis.conf, you can go ahead and restart the Redis server.
In the demo repo, I’ve included the data set we cobbled together by using the redisgraph-bulk-loader script to load the following from CSV into RedisGraph. This dataset only includes the mammals in the animal kingdom, due to the lower quality of data available for animals outside of mammals (Non-mammals species rarely have good Wikipedia descriptions).
Here is how you load the data:
$ cd redisgraph-bulk-loader/
$ python3 bulk_insert.py MAMMALS -q -n /path/to/demo/dataload/Class.csv -n /path/to/demo/dataload/Family.csv -n /path/to/demo/dataload/Genus.csv -n /path/to/demo/dataload/Order.csv -n /path/to/demo/dataload/Species.csv -r /path/to/demo/dataload/IN_CLASS.csv -r /path/to/demo/dataload/IN_FAMILY.csv -r /path/to/demo/dataload/IN_GENUS.csv -r /path/to/demo/dataload/IN_ORDER.csv -a yourpassword
1 nodes created with label 'Class'
157 nodes created with label 'Family'
1272 nodes created with label 'Genus'
29 nodes created with label 'Order'
5616 nodes created with label 'Species'
29 relations created for type 'IN_CLASS'
1272 relations created for type 'IN_FAMILY'
5616 relations created for type 'IN_GENUS'
157 relations created for type 'IN_ORDER'
Construction of graph 'MAMMALS' complete: 7075 nodes created, 7074 relations created in 0.443749 seconds
$ redis-cli -a yourpassword GRAPH.QUERY MAMMALS "CALL db.idx.fulltext.createNodeIndex('Species','description')"
Warning: Using a password with '-a' or '-u' option on the command line interface may not be safe.
1) (empty list or set)
2) (empty list or set)
3) 1) "Query internal execution time: 324.970000 milliseconds"
(gist: https://gist.github.com/stockholmux/0727a4a784a46f8cb9e8329d393a513a)
At this point, the key MAMMALS contains our entire graph. A couple of important notes:
- The -q switch on bulk_insert.py is very important because it allows for smart quoting when reading in the CSV.
- The redis-cli call does a bulk indexing of all the nodes at one time, so, in effect, it’s ingesting over 7,000 documents for full-text search.
Now, let’s get a UI up and running. As with practically any Node.js application, we start with npm install. This might take a few seconds, as we’re managing not only the server-side files for Node.js, but also the front-end Vue.js components. If you haven’t spent much time in the front-end Javascript world recently, it’s no longer a FTP and HTML file affair. Modern front-enders do take their tooling seriously, so we’ll need Vue CLI installed (I suggest following the Vue CLI getting started guide).
After you’ve got your front-end tooling in place, let’s float back to npm and build the front end by running:
$ npm run build
This will create the dist directory that we’ll serve all our front-end files from. Now that we have our data in Redis, our front-end files are ready to serve, so we can kick on the server:
$ node server.js -p 6379 -a yourpassword -h yourhostOrlocalhost
Let’s pause for a moment and cover a few points about this server we just turned on. It’s built on Express.js and primarily uses websockets to communicate back and forth. I also integrated the Visible Engine debugging tool, which allows you to see commands being executed in a separate browser window. You can play with that demo by pointing your browser at http://localhost:4444.
All in all, it’s remarkably short for what it does – only 75 lines of code. Our solution doesn’t need to be that long because all we’re really doing is accepting websocket connections, running Redis commands based on the passed messages, and passing those messages back with the results. Redis(Graph) is doing all the hard work here. Let’s take a look at the commands we’re executing.
To search for keywords, we run this command:
> GRAPH.QUERY MAMMALS "CALL db.idx.fulltext.queryNodes('Species','cat house pet')"
This is pretty unsophisticated. Our key is MAMMALS and we using a special Cypher syntax CALL to a specific function, which passes in the label of the nodes we’re looking for and finally the actual search string. You can pass in valid RediSearch queries, but keep in mind that this is only full-text search at the moment, so don’t use geospatial, tag or numeric clauses.
Once we’ve identified the two animals we’re comparing, we use a straight Cypher query:
> GRAPH.QUERY MAMMALS "MATCH (s:Species)-[*]->(x)<-[*]-(c:Species) WHERE c.fullname = 'Felis catus' AND s.fullname = 'Balaenoptera borealis' RETURN x.name, labels(x) LIMIT 1"
In the server.js file, these queries are represented as Javascript template strings. Nothing is hidden, it’s simply user input through string interpolation to a query. However, if deploying something like this in production, you’d want to be careful with accepting user input.
If you plan to tinker with the front-end code, make sure you edit the /src directory, not /dist. After editing, you’ll need to run npm run build again or use the development server (npm run serve) that auto-compiles changes to front-end code and serves it up on another port. Again, not much going on here, it’s a pretty standard Vue.js and Bootstrap application. The only really relevant files are /src/App.js, /src/components/panels.vue and /src/components/search.vue.
That’s it. A simple demo to show off a powerful feature that integrates two distinct models – graph and full-text search. I encourage you to play with the demo and sub in your own datasets.
Get started with Redis today
Speak to a Redis expert and learn more about enterprise-grade Redis today.
